

The goal of wideRhino is to enable the construction of
canonical variate analysis (CVA) biplots for high-dimensional data
settings, specifically where the number of variables (\(p\)) exceeds the number of observations
(\(n\)). The package addresses the
singularity limitation of the within-group scatter matrix by leveraging
the generalised singular value decomposition (GSVD).
You can install the development version of wideRhino from GitHub with:
# install.packages("pak")
pak::pak("RaeesaGaney91/wideRhino")When \(p < n\), then the CVA-GSVD
biplot will result to the standard CVA biplot. Here is an example using
the penguins data:
library(wideRhino)
Penguins <- datasets::penguins[stats::complete.cases(penguins),]
CVAgsvd(X=Penguins[,3:6],group = Penguins[,1]) |>
CVAbiplot(group.col=c("blue","purple","forestgreen"))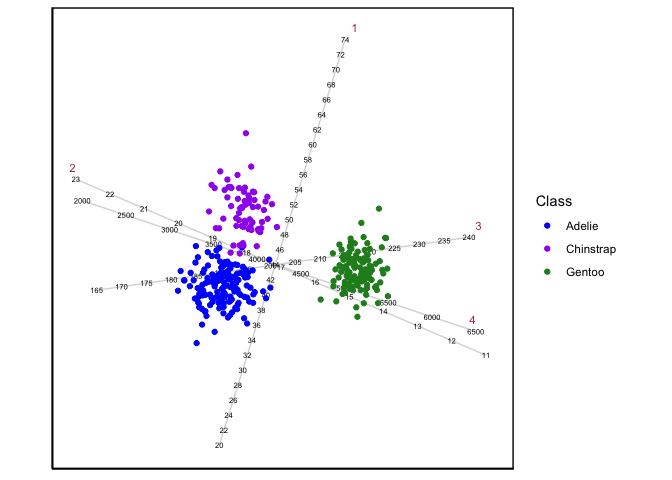
When \(p > n\), then the standard CVA biplot will not work due to the singularity of the within-scatter matrix, and this is when the GSVD becomes useful. Using a simulated data set with 3 groups, 100 observations and 300 variables, a CVA-GSVD biplot can be constructed:
data(sim_data)
CVAgsvd(X=sim_data[,2:301],group = sim_data[,1]) |>
CVAbiplot(group.col=c("tan1","darkcyan","darkslateblue"),which.var = 1:10,zoom.out=80)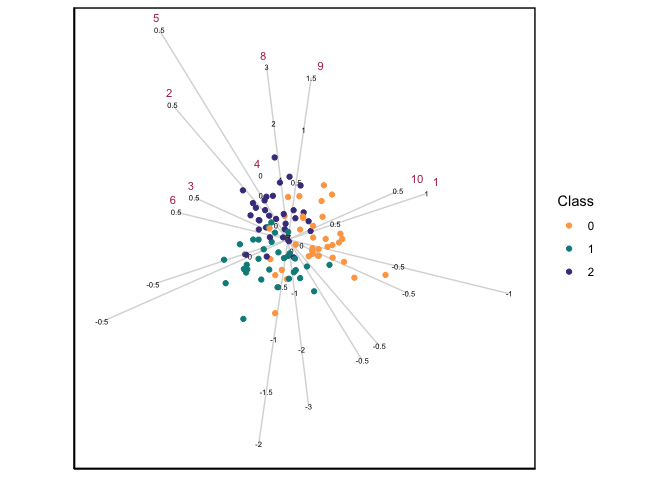
The name wideRhino is inspired by the white rhinoceros,
a species distinguished by its wide mouth and short legs. This physical
structure reflects the statistical characteristics of the data the
package is designed for: wide data with a large number of variables
(\(p\)) and a small number of
observations (\(n\)) — a setting often
described as “large \(p\), small \(n\)”.
Just as the white rhino’s wide frame is well-adapted to its environment, wideRhino is purpose-built for the challenges of high-dimensional multivariate analysis. By leveraging the generalised singular value decomposition (GSVD), it allows users to construct canonical variate analysis (CVA) biplots even when classical assumptions break down.