

{shapviz} provides typical SHAP plots:
sv_importance(): Importance plot (bar/beeswarm).sv_dependence() and sv_dependence2D():
Dependence plots to study feature effects and interactions.sv_interaction(): Interaction plot (beeswarm/bar).sv_waterfall(): Waterfall plot to study single or
average predictions.sv_force(): Force plot as alternative to waterfall
plot.SHAP and feature values are stored in a “shapviz” object that is built from:
We use {patchwork} to glue together multiple plots with (potentially) inconsistent x and/or color scale.
# From CRAN
install.packages("shapviz")
# Or the newest version from GitHub:
# install.packages("devtools")
devtools::install_github("ModelOriented/shapviz")Shiny diamonds… let’s use XGBoost to model their prices by the four “C” variables:
library(shapviz)
library(ggplot2)
library(xgboost)
set.seed(1)
xvars <- c("log_carat", "cut", "color", "clarity")
X <- diamonds |>
transform(log_carat = log(carat)) |>
subset(select = xvars)
# Fit (untuned) model
fit <- xgb.train(
params = list(learning_rate = 0.1),
data = xgb.DMatrix(data.matrix(X), label = log(diamonds$price)),
nrounds = 65
)
# SHAP analysis: X can even contain factors
X_explain <- X[sample(nrow(X), 2000), ]
shp <- shapviz(fit, X_pred = data.matrix(X_explain), X = X_explain)
sv_importance(shp, show_numbers = TRUE)
sv_importance(shp, kind = "bee")
sv_dependence(shp, v = xvars, share_y = TRUE)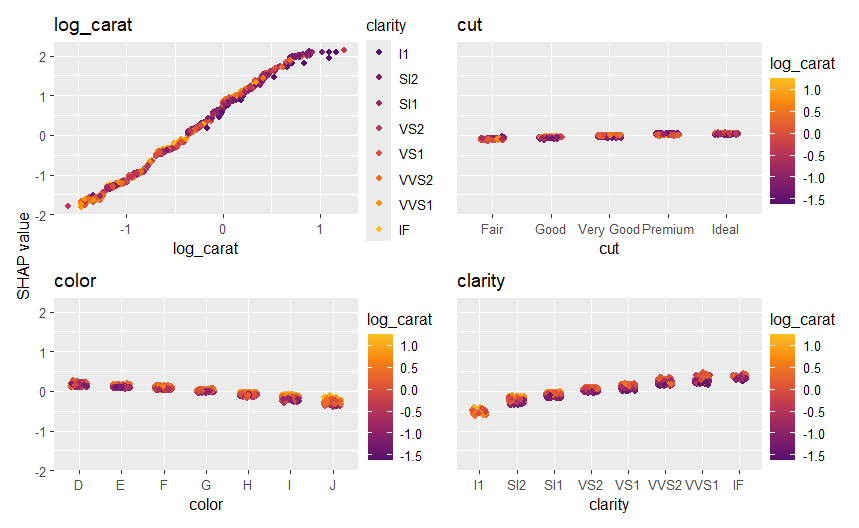
Decompositions of individual predictions can be visualized as waterfall or force plot:
sv_waterfall(shp, row_id = 2) +
ggtitle("Waterfall plot for second prediction")
sv_force(shp, row_id = 2) +
ggtitle("Force plot for second prediction")Check-out the vignettes for topics like:
[1] Scott M. Lundberg and Su-In Lee. A Unified Approach to Interpreting Model Predictions. Advances in Neural Information Processing Systems 30 (2017).